A Blood-Based Molecular Clock for Biological Age Estimation
Ersilia Paparazzo, Silvana Geracitano, Denise Bartolomeo, Mirella Aurora Aceto, Patrizia D’Aquila, Dina Bellizzi, Giuseppe Passarino, Alberto Montesanto, Department of Biology, Ecology and Earth Sciences, University of Calabria, Rende; Vincenzo Lagani, Biological and Environmental Sciences and Engineering Division (BESE), King Abdullah University of Science and Technology KAUST, Thuwal, Institute of Chemical Biology, Ilia State University, 0162 Tbilisi, SDAIA-KAUST Center of Excellence in Data Science and Artificial Intelligence, Thuwal; Luigi Citrigno, National Research Council (CNR)—Institute for Biomedical Research and Innovation—(IRIB), Mangone
Digital Library: https://doi.org/10.3390/cells12010032
Abstract
In the last decade, extensive efforts have been made to identify biomarkers of biological age. DNA methylation levels of ELOVL fatty acid elongase 2 (ELOVL2) and the signal joint T-cell receptor rearrangement excision circles (sjTRECs) represent the most promising candidates. Although these two non-redundant biomarkers echo important biological aspects of the ageing process in humans, a well-validated molecular clock exploiting these powerful candidates has not yet been formulated. The present study aimed to develop a more accurate molecular clock in a sample of 194 Italian individuals by re-analyzing the previously obtained EVOLV2 methylation data together with the amount of sjTRECs in the same blood samples. The proposed model showed a high prediction accuracy both in younger individuals with an error of about 2.5 years and in older subjects where a relatively low error was observed if compared with those reported in previously published studies. In conclusion, an easy, cost-effective and reliable model to measure the individual rate and the quality of aging in human population has been proposed. Further studies are required to validate the model and to extend its use in an applicative context.
Methods:
The dataset in this study includes 194 unrelated individuals (90 men and 104 women) with a mean age of 63.5 years (2002 to 2007 time interval). Peripheral blood samples were collected to extract DNA and genomic DNA.
Methylation analysis of nine consecutive CpGs in the ELOVL2 gene region was performed by bisulfite pyrosequencing. Real-time PCR conditions and primers for quantification of sjTREC were carried out according to previously published technique. Pearson correlation coefficient and linear regression were used for assessing the univariate association between biological markers and age.
Results
Re-assessing the DNA methylation markers of ELOLV2 gene together with the analysis of sjTREC content in the blood, showed that the sjTREC levels and DNA methylation of the ELOVL2 gene are very useful markers for age prediction. In fact, the SVR model developed with JADBio, including these two non-redundant markers showed high prediction accuracy with a prediction error of about 4.4 years, dropping down to lower that 2.5 years in younger individuals.
Conclusions: The data reported in this study may be of interest, because an easy, cost-effective and reliable model was proposed that might represent a new tool to measure the individual rate and the quality of aging in geriatric ages.
How was JADBio used?
Development of a Blood-Based Molecular Clock
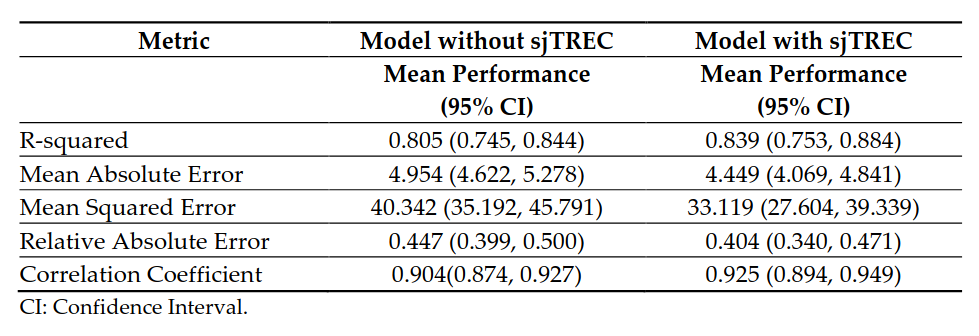
Ιn order to obtain more accurate age prediction models, a machine learning approach was adopted, using JADBio (v1.3.32). This approach produced two final best models, one using the average methylation value of the ELOVL2 promoter region as only predictor, and the coupling ELOVL2 with sjTREC. Both models were trained using Support Vector Regression Machines (SVR) with Polynomial Kernel as the best performing models. Their predictive performances are reported in Table 3. Further details on the two models are available on the JADBio platform (links available below).
Prediction accuracies of the SVR models obtained in JADBio using the average methylation value of the ELOVL2 promoter region alone or in combination with sjTREC content (Table 3)
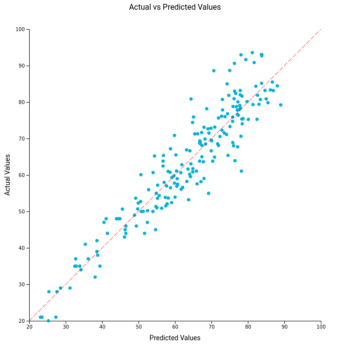
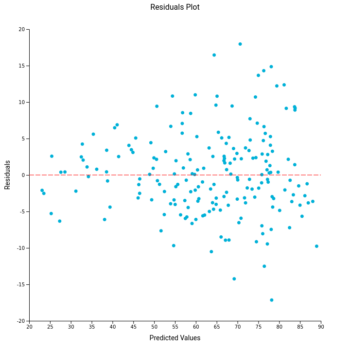
Age prediction from blood-derived DNA samples. (Left) Linear regression of the relationships between human individual age and age predicted using the molecular clock in which ELOVL2 was used in combination with sjTREC (dotted lines correspond to 95% prediction interval). (Right) Deviation of DNA methylation (predicted) age and chronological age (residuals) against the predicted age. The deviation in older individuals (>60 years old) was higher compared to younger ones. Linear and LOESS regression lines are reported in dashed red on the left and right panel, respectively (Figure 3)
Read more at MDPI – Cells: A Blood-Based Molecular Clock for Biological Age Estimation
OTHER
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Stay connected to get our news first!
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Join the JADai Community!
Sign up with a FREE Basic plan! Be part of a growing community of AutoML enthusiasts
GET STARTED