Urine and Fecal ¹H-NMR Metabolomes Differ Significantly between Pre-Term and Full-Term Born Physically Fit Healthy Adult Males
Leon Deutsch, Simona Opara, Department of Animal Science, Biotechnical Faculty, University of Ljubljana; Tadej Debevec, Faculty of Sports, University of Ljubljana, Department of Automation, Biocybernetics and Robotics, Jožef Stefan Institute, Ljubljana; Gregoire P. Millet, Institute of Sport Sciences, University of Lausanne; Damjan Osredkar, Department of Pediatric Neurology, University Children’s Hospital, University Medical Centre Ljubljana, Faculty of Medicine, University of Ljubljana; Robert Šket, Institute for Special Laboratory Diagnostics, University Children’s Hospital, University Medical Centre Ljubljana; Boštjan Murovec, Faculty of Electrical Engineering, University of Ljubljana; Minca Mramor, Department of Infectious Diseases, University Medical Centre Ljubljana; Janez Plavec, National Institute of Chemistry, NMR Center, Ljubljana; Blaz Stres, Department of Animal Science, Biotechnical Faculty, University of Ljubljana, Department of Automation, Biocybernetics and Robotics, Jožef Stefan Institute, Ljubljana, Institute of Sanitary Engineering, Faculty of Civil and Geodetic Engineering, University of Ljubljana
Digital Library: https://www.mdpi.com/2218-1989/12/6/536
Abstract
Preterm birth (before 37 weeks gestation) accounts for ~10% of births worldwide and remains one of the leading causes of death in children under 5 years of age. Preterm born adults have been consistently shown to be at an increased risk for chronic disorders including cardiovascular, endocrine/metabolic, respiratory, renal, neurologic, and psychiatric disorders that result in increased death risk. Oxidative stress was shown to be an important risk factor for hypertension, metabolic syndrome and lung disease (reduced pulmonary function, long-term obstructive pulmonary disease, respiratory infections, and sleep disturbances).
Methods: The aim of this study was to explore the differences between preterm and full-term male participants’ levels of urine and fecal proton nuclear magnetic resonance (¹H-NMR) metabolomes, during rest and exercise in normoxia and hypoxia and to assess general differences in human gut-microbiomes through metagenomics at the level of taxonomy, diversity, functional genes, enzymatic reactions, metabolic pathways and predicted gut metabolites.
Results
Significant differences existed between the two groups based on the analysis of ¹H-NMR urine and fecal metabolomes and their respective metabolic pathways, enabling the elucidation of a complex set of microbiome related metabolic biomarkers, supporting the idea of distinct host-microbiome interactions between the two groups and enabling the efficient classification of samples; however, this could not be directed to specific taxonomic characteristics.
Conclusions: The results presented in this study constitute the first report on the differences in the urine and fecal metabolomes between preterm and full-term groups of physically fit healthy young males. Clear differences were identified in the urine and fecal metabolomes next to the metabolic pathways, suggesting that systemic differences between the two groups affect the metabolism of the host as well as intestinal tract parameters and that of the underlying microbiome (supplementary material, Figure S13).
How was JADBio used?
Significant differences in urine and fecal 1H-NMR metabolomes between the preterm and full-term groups prompted led to exploring whether significant differences exist at the level of the human gut microbiome. Fecal samples collected during the PreTerm project were used for shotgun sequencing and after preprocessing and analysis, 393,442 variables were considered in this search for differences between the preterm and full-term groups. In total, 853 taxonomic units (kingdoms, phyla, clades, orders, families, genera, and species), 30 diversity calculators, 198,305 gene families, 183,200 enzymatic reactions, 10,974 metabolic pathways, and 80 metabolites present in the human gut microbiota were identified and analyzed. Each dataset corresponding to a layer of information was analyzed separately using JADBio extensive machine learning modeling.
JADBIO AutoML platform was used to search for biomarkers. Personal analytic biases and methodological statistical errors were eliminated from the analysis by autonomously exploring different settings in the modeling steps, resulting in more convincing discovered features to distinguish between different groups. JADBIO was used to model different dataset choices in addition to the features observed in samples of all groups from different projects by splitting the total data into a training set and a test set in a 70:30 ratio. The training set was used to train the model and the test set was used to evaluate the model [39,56].
To assess the classification of the model, a receiver-operating characteristic curve (ROC curve) was constructed for all studied groups, plotting the true-positive rate (sensitivity) against the false-positive rate (1-specificity). Individual conditional expectation plots (ICE) showed the nature of the contribution of each feature characteristic to the model. All obtained models can be run locally using a Java executor (supplementary materials).
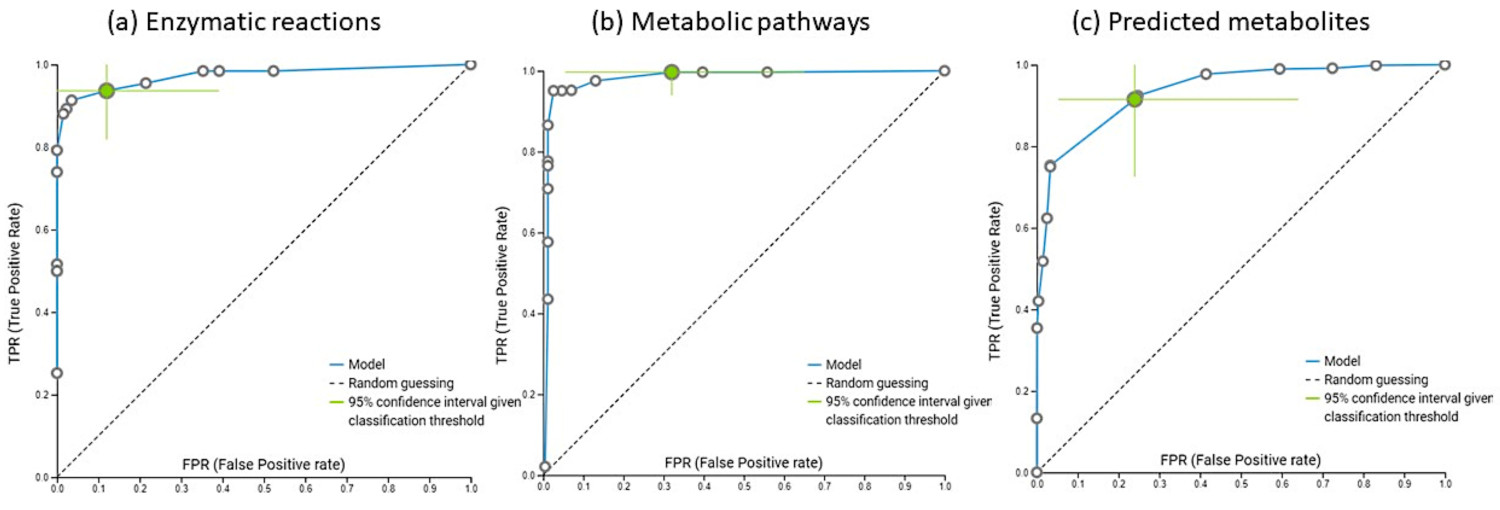
ROC curves of obtained JADBio models based on the enzymatic reactions (a), metabolic pathways (b) and relaxation network predicted metabolites (c) produced by in-house implementation of bioBakery3. (Figure 4)
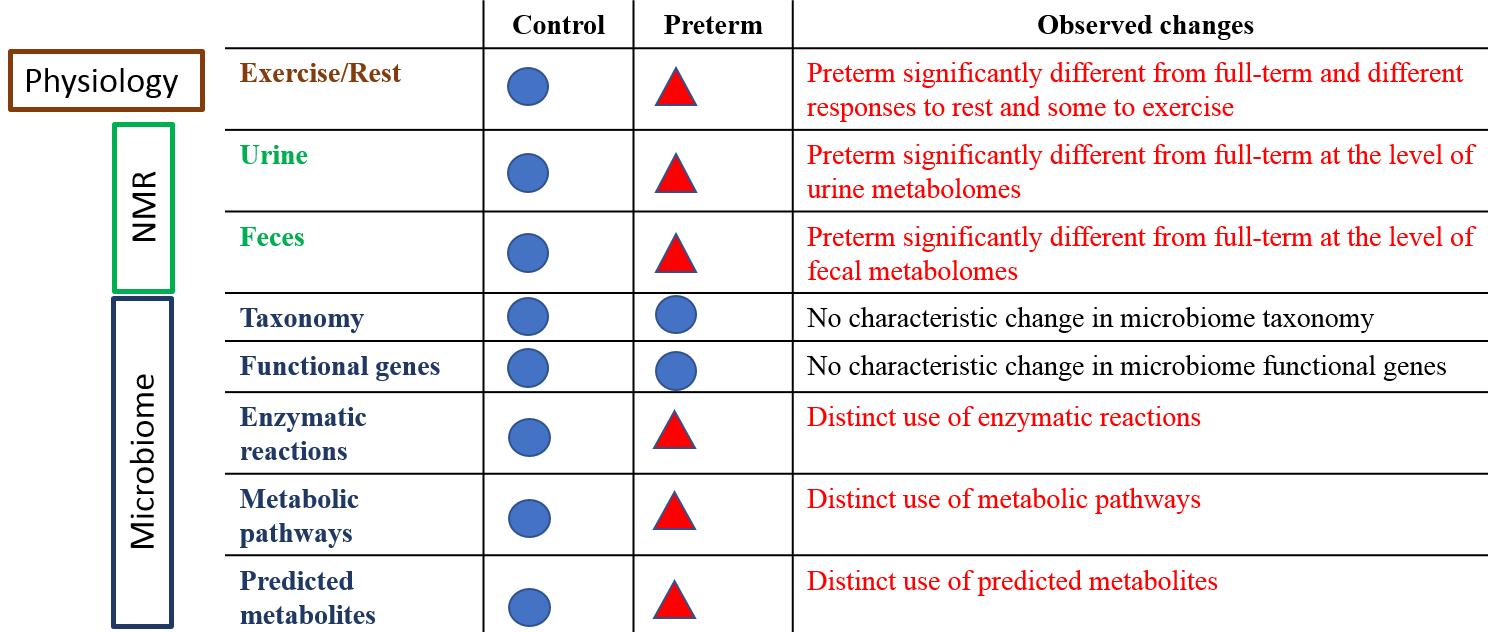
A summary of observed changes at various information levels showing that significant differences exist between the preterm and full-term adult urine metabolomes, fecal metabolomes and microbial metabolic reactions and pathways. Taken together, these results show that host and its microbiome behave measurably different in healthy physically fit young males in comparison to matched full-term controls. (Supplementary materials, Figure S13)
Supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/metabo12060536/s1, ESM1: ESM 1; ESM 2: Metabolites identified in urinary samples; ESM 3: Metabolites identified in fecal samples; ESM 4: Models with instructions for local running.
Read more at MDPI – metabolites: Urine and Fecal 1H-NMR Metabolomes Differ Significantly between Pre-Term and Full-Term Born Physically Fit Healthy Adult Males
OTHER
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Stay connected to get our news first!
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Join the JADai Community!
Sign up with a FREE Basic plan! Be part of a growing community of AutoML enthusiasts
GET STARTED



