Tissue-Specific Methylation Biosignatures for Monitoring Diseases: An In Silico Approach
Makrina Karaglani, Maria Panagopoulou, Paraskevi Apalaki, Theodosis Theodosiou, Laboratory of Pharmacology, Department of Medicine, Democritus University of Thrace; Ismini Baltsavia, Ioannis Iliopoulos, School of Medicine, Department of Basic Sciences, University of Crete; Ioannis Tsamardinos, JADBio Gnosis DA, Science and Technology Park of Crete, Department of Computer Science, University of Crete, Institute of Applied and Computational Mathematics, Foundation for Research and Technology-Hellas; Ekaterini Chatzaki, Laboratory of Pharmacology, Department of Medicine, Democritus University of Thrace, Institute of Agri-Food and Life Sciences, Hellenic Mediterranean University Research Centre
Digital Library: https://www.mdpi.com/1422-0067/23/6/2959
Abstract
Tissue-specific gene methylation events are key to the pathogenesis of several diseases and can be utilized for diagnosis and monitoring. In this paper, an in silico pipeline was established to analyze high-throughput methylome datasets to identify specific methylation fingerprints in three pathological entities of major burden, i.e., breast cancer (BrCa), osteoarthritis (OA) and diabetes mellitus (DM).
Methods: Differential methylation analysis was conducted to compare tissues/cells related to the pathology and different types of healthy tissues, revealing Differentially Methylated Genes (DMGs). Highly performing and low feature number biosignatures were built with automated machine learning, including: (1) a five-gene biosignature discriminating BrCa tissue from healthy tissues (AUC 0.987 and precision 0.987), (2) three equivalent OA cartilage-specific biosignatures containing four genes each (AUC 0.978 and precision 0.986) and (3) a four-gene pancreatic β-cell-specific biosignature (AUC 0.984 and precision 0.995). Next, the BrCa biosignature was validated using an independent ccfDNA dataset showing an AUC and precision of 1.000, verifying the biosignature’s applicability in liquid biopsy.
Results
Functional and protein interaction prediction analysis revealed that most DMGs identified are involved in pathways known to be related to the studied diseases or pointed to new ones.
Conclusions: Overall, used data-driven approach contributes to the maximum exploitation of high-throughput methylome readings, helping to establish specific disease profiles to be applied in clinical practice and to understand human pathology.
How was JADBio used?
BrCa-Specific Methylation Biosignature through AutoML
β-values produced by RnBeads Differential Methylation Analysis were analyzed using JADBio in order to construct an accurate model specific for tracing BrCa. The original dataset (218 BrCa tissues and 193 healthy tissues) was automatically and randomly split into a training dataset of 151 BrCa and 131 healthy tissues and a validation dataset of 66 BrCa and 55 healthy tissues. Analysis of the training dataset of 29,703 gene array features produced one signature containing 5 features via a support vector machines (SVM) algorithm (https://app.jadbio.com/share/4fd50c38-d0a1-4f28-96c9-480b29b4a3e2, accessed on 1 October 2021). Three of them were protein-coding genes, namely, CCDC181, HIST2H3PS2 and CFTR, and two were RNA genes, namely, RUVBL1-AS1 and AL161908.1 (Table 1). All genes presented increased methylation in BrCa in relation to healthy tissues/cells. In discriminating BrCa against healthy tissues, this signature reached an area under the curve (AUC) of 0.987 (0.963–1.000) and an average precision of 0.987 (0.955–1.000) (Figure 2A). Upon validation in the test dataset, the model showed an AUC and an average precision of 0.995 (Figure 2A), verifying the model’s performance metrics. The performance and inspection results are depicted in Figure 2B–D.
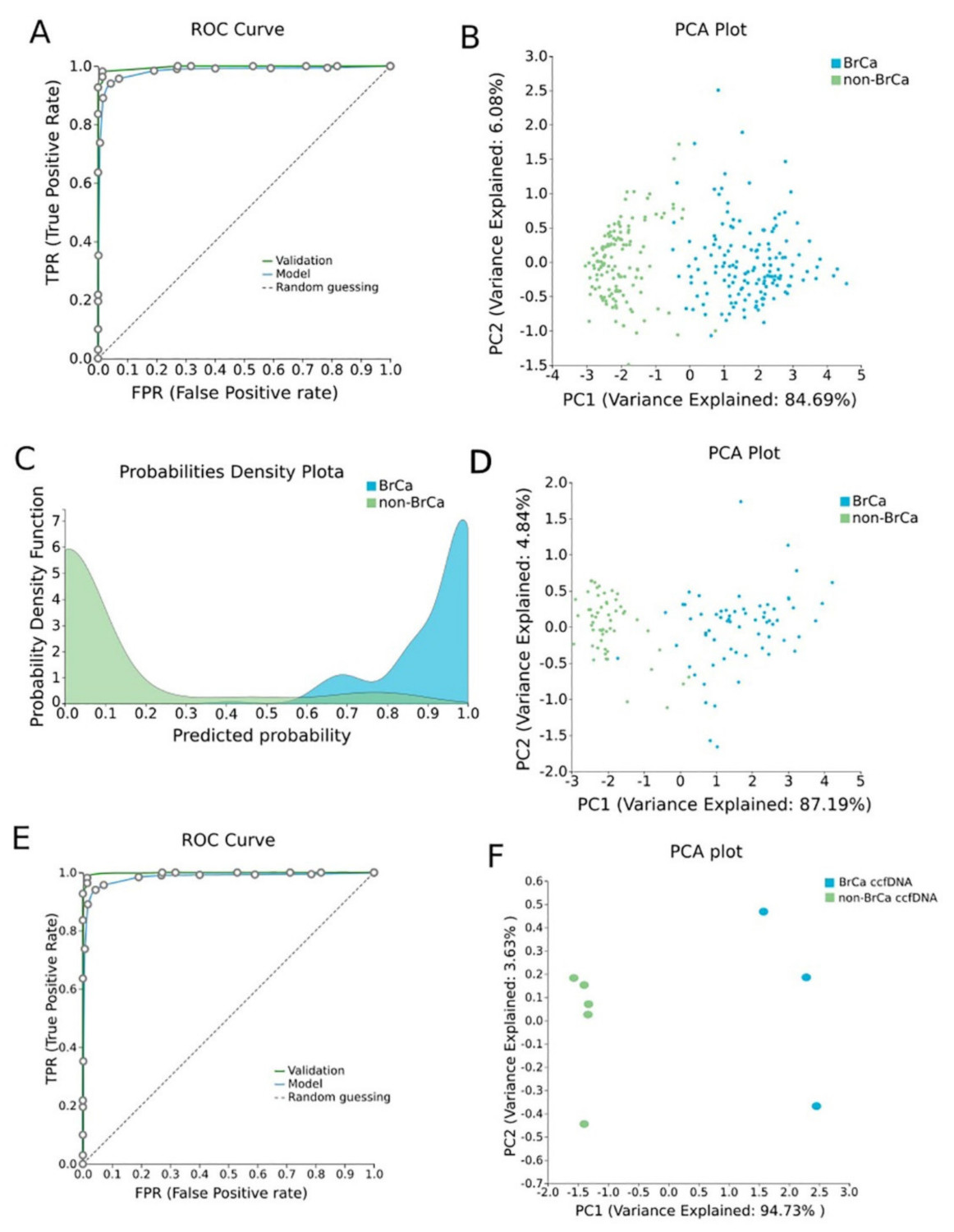
BrCa-specific methylation biosignature built using AutoML (Figure 2)
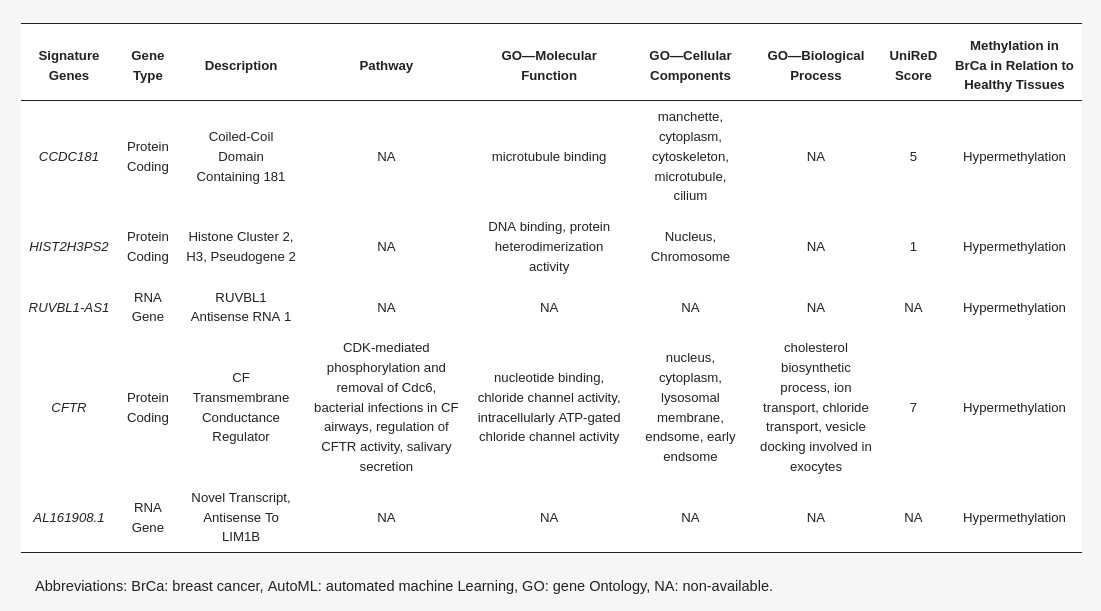
Differentially methylated genes selected in the BrCa-specific signature built using AutoML analysis (Table 1)
To validate the discrimination performance of the BrCa-specific five-feature biosignature on ccfDNA and its applicability to liquid biopsy, we applied it to an external independent dataset of three BrCa ccfDNA samples and five ccfDNA samples from age-matched healthy women. The analysis revealed the model’s AUC and an average precision of 1.000 (Figure 2E,F).
OA Specific Methylation Biosignature through AutoML
In order to construct a specific model for OA, β-values were uploaded to JADBio. The original dataset (151 OA tissues and 216 healthy tissues) was automatically and randomly split into a training dataset of 108 OA and 144 healthy tissues and a validation dataset of 43 OA and 65 healthy tissues. An analysis of the training dataset of 29,585 gene array features produced three equivalent signatures containing 4 features each via a classification random forests algorithm (https://app.jadbio.com/share/8a2ac85c-10b4-4d65-9a70-ae9377df7878, accessed on 1 October 2021). Two of them were protein-coding genes, namely CASD1 and STOML1, two were lncRNA genes, namely, LINC01350 and RP11-272L13.3, and one was an RNA gene, namely, CARMAL. The last was the RP11-515E23.2 gene (Table 2). Common features between models were RP11-515E23.2, LINC01350 and CASD1. All genes showed the down-regulation of methylation in OA cartilage in relation to healthy tissues. In discriminating OA against healthy tissues, signatures reached an AUC of 0.978 (0.942–1.000) and average precision of 0.986 (0.962–1.000) (Figure 4A). Upon validation, the model showed an AUC of 0.990–0.995 and an average precision of 0.994–0.997 (Figure 4A), verifying the stability and accuracy of its estimation. Performance validation and inspection are depicted in Figure 4B,C.
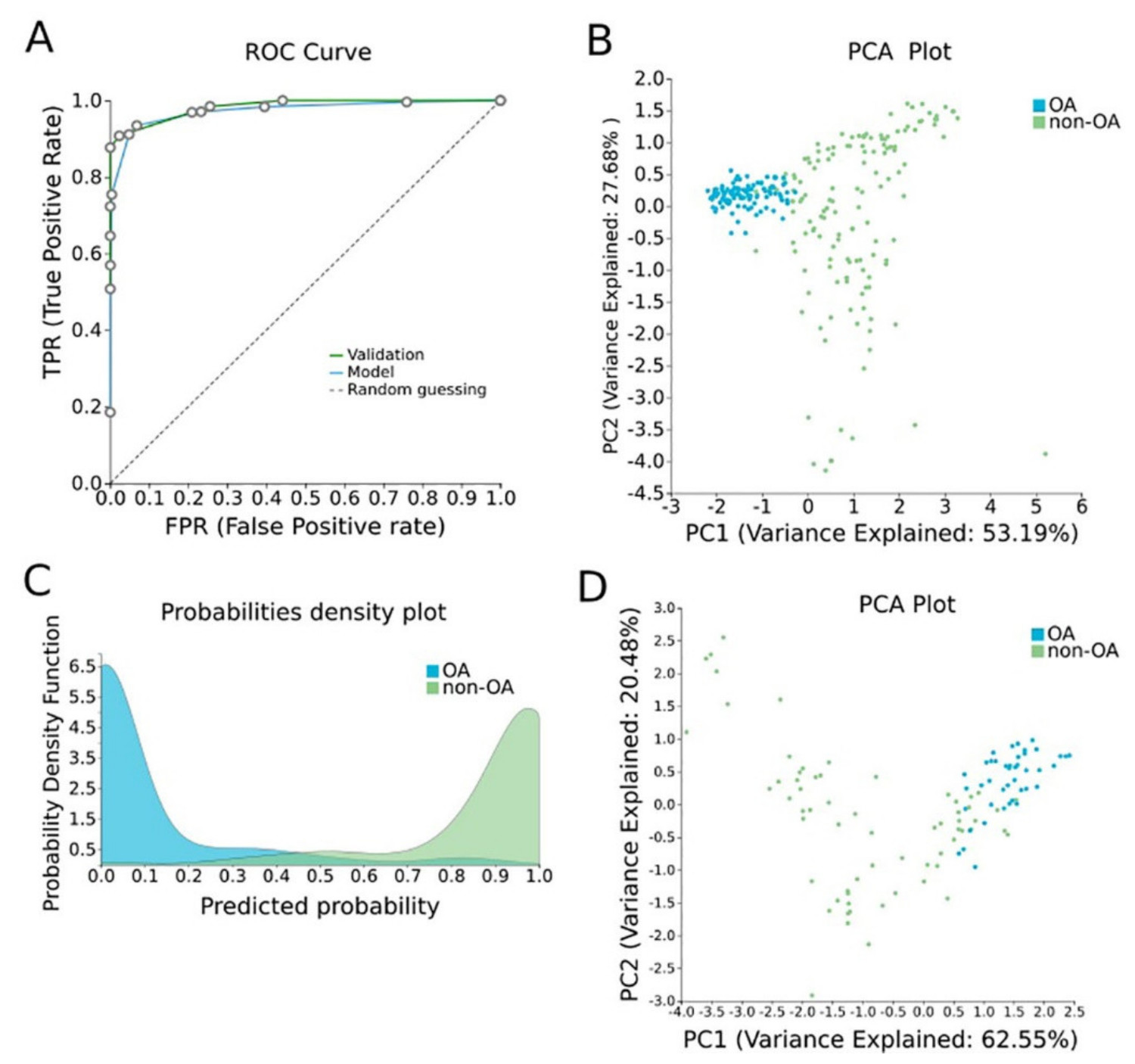
OA-specific methylation biosignature built using AutoML (Figure 4)
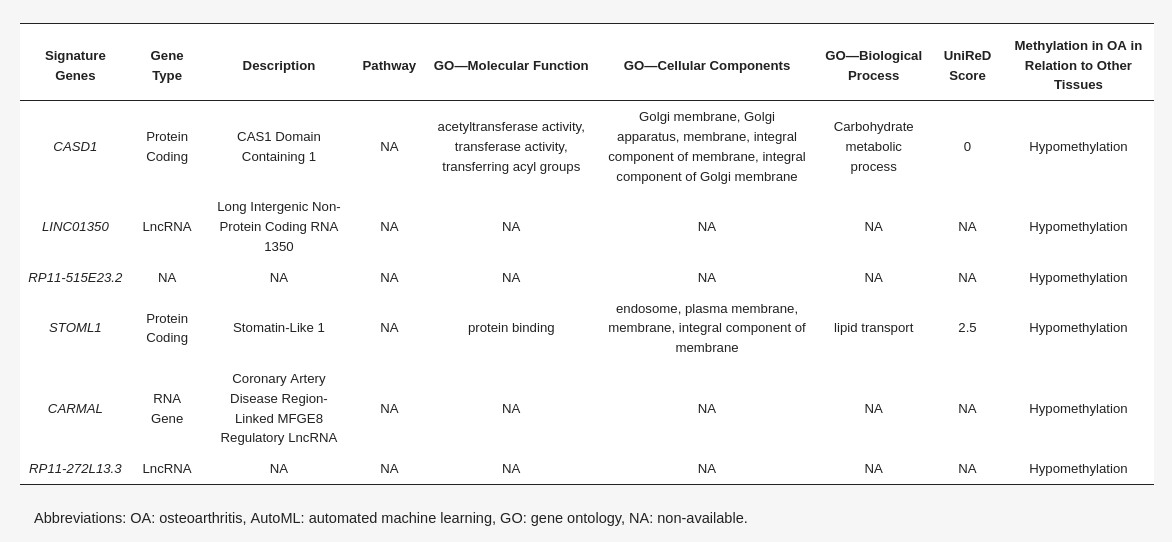
Differentially methylated genes selected in the OA cartilage-specific signature built using AutoML analysis (Table 2)
Pancreatic β-Cell Specific Methylation Biosignature Using AutoML
To construct a pancreatic β-cell-specific methylation biosignature, methylome β-values of 3 β-cell samples and 28 other tissue/cell samples were analyzed through JADBio. From the 28,021 CG feature dataset, AutoML analysis produce a biosignature containing 4 features via a support vector machine algorithm (https://app.jadbio.com/share/7ebbc7c3-b861-41af-8a39-88202756d609, accesed on 1 October 2021). Two of them were protein-coding genes, namely, TXNRD3 and LENG8, one was a snoRNA gene, namely, SCARNA6, and one an LncRNA gene, namely, AC008741.1 (Table 3). All genes showed decreased methylation in pancreatic β-cells in relation to other tissues/cells. The signature’s performance in discriminating β-cells reached an AUC of 0.984 (0.909–1.000) and an average precision of 0.995 (0.975–1.000) (Figure 6A). The model’s performance and inspection are depicted in Figure 6B,C.
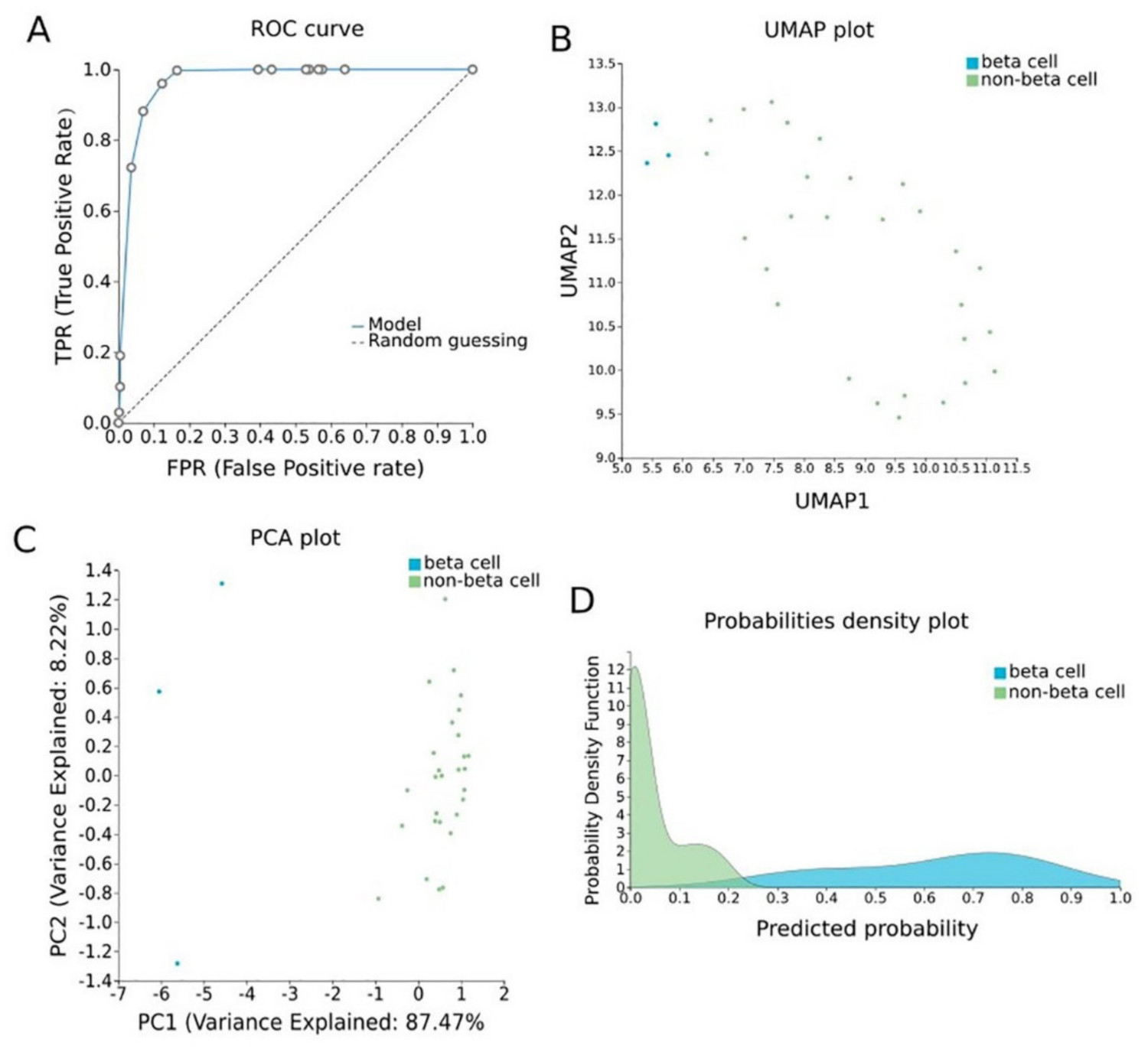
Pancreatic β-cell-specific methylation biosignature built using AutoML (Figure 6)
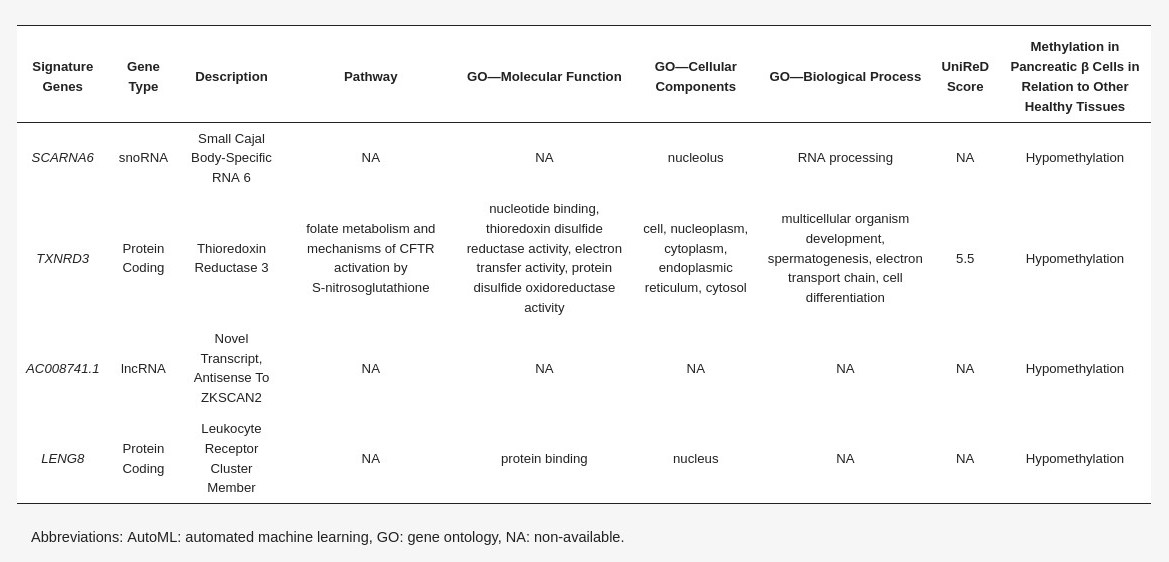
Differentially methylated genes selected in the pancreatic β-cell-specific signature built using AutoML analysis comparing methylomes of β-cells and other healthy tissues (Table 3)
Using AutoML, the research team was able to construct a five-gene signature exhibiting a high AUC of 0.987 and a precision of 0.987 when discriminating BrCa against healthy tissues.
Most importantly, AutoML analysis delivered three equivalent OA cartilage-specific biosignatures with high performance (AUC of 0.978 and precision of 0.986) containing four features each.
Specific methylation patterns of pancreatic β-cells would be of great value in the early detection and monitoring of pancreatic cell loss during diabetes. A highly performing biosignature (AUC of 0.984 and precision of 0.995) was developed through AutoML analysis. The biosignature contained two protein-coding genes, namely, TXNRD3 and LENG8, one snoRNA gene and one LncRNA gene. Only TXNRD3 was found to be associated with diabetes-related pathways, reaching a score of 5.5 in machine learning-aided text mining.
In order to build methylation-based biosignatures, AutoML was employed, for the first time, using JADBio. This approach presents two advantages of major significance for further developments in biomarker discovery: (1) It has high-performing classifiers with low feature numbers via feature selection, i.e., automatic calculations for identifying the minimum feature number within a dataset of some thousands of features that retain the maximum classifying power. Reducing the dimensions of a signature is a great advantage in terms of translatability to cost-effective assays with less technical requirements for multiplexing, moving from the multi-dimensional omics results to simpler classifiers. Upon prospective clinical validation, these signatures can offer feasible solutions for laboratory tests that could be realized in any standardly equipped diagnostic lab. (2) JADBio has been shown to shield against typical methodological pitfalls in data analysis that lead to overfitting and overestimating performance and, therefore, to misleading results. This is again confirmed here, as the AUC of the biosignatures built did not fall significantly when tested in the validation sub-datasets or in independent cohorts, adding credibility to this approach, showing that it can deliver mature solutions for clinical development.
OTHER
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Stay connected to get our news first!
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Join the JADai Community!
Sign up with a FREE Basic plan! Be part of a growing community of AutoML enthusiasts
GET STARTED



