Multi-omics Characterization of Left-Right Colorectal Cancer
John Marshall, Takayuki Yoshino, Sun Young Rha, David N. Church, Anelisa Kruschewsky Coutinho, Carlos Alberto Sampaio-Filho, David James Gallagher, Jesús García-Foncillas, Silvia von der Heyde, Hartmut Juhl, Jonathan Woodsmith, David J. Kerr
Journal of Clinical ONCOLOGY: https://ascopubs.org/doi/abs/10.1200/JCO.2021.39.15_suppl.3542
Abstract
Right (R) vs left (L) sided colorectal cancers are clinically distinguishable based on prognosis and response to certain therapies, but as of yet, limited data have emerged to explain these differences. The science of molecular testing has evolved rapidly. Enabled by improved technologies and computing power, it is now feasible to obtain to systematic multi-omic datasets covering DNA, RNA, proteins, phospho-proteins and metabolomics on large numbers of patients. Multi-omic analysis can further define disease specific subgroups but pre-analytic quality of the tissues (ischemia time) and comparison to normal tissue controls is paramount to optimize results. Methods: Following informed consent, 450 colorectal cancer primary tumors and paired normal tissues were collected following an SOP to minimize ischemia time, and were analyzed using comprehensive genomics, transcriptomics, proteomics, phosphoproteomics, morphology and annual clinical information. Right (C18.0,2,3) and left (C18.6,7) CRC tumors, normal tissue were compared using machine learning tools to unravel the molecular mechanisms that underpin these clinically distinguishable phenotypes as well as correlating with known genomic metrics such MSI and KRAS mutation status. Results: Through leveraging the tumor and paired normal patient samples, systematic differences between left and right tumor samples were observed including specific molecular events associated with these anatomical differences. The detailed results will be presented at the meeting. Conclusions: Progress in precision medicine requires the inclusion of multi-omics which in turn requires changes to our current SOPs of tissue collection. The ability to define molecular distinctions such as between R and L colon cancer will permit the rapid discovery of clinically useful prognostic and predictive markers, dramatically adding to our fundamental understanding to colon cancer biology. Future work will focus on the discovery of novel targets and signatures, creating innovative tools that depict multi-omic results for clinicians. Read more in the Journal of Clinical ONCOLOGY: Multi-omics characterization of left-right colorectal cancer
How was JADBio used?
Supervised machine learning approach, using JADBio was used to classify left and right CRC based on the fold change in values between tumor and adjacent normal. The ROC and AUC (AUC = 0.995) show that the model can almost perfectly classify left and right- based tumors (Figure 6a).
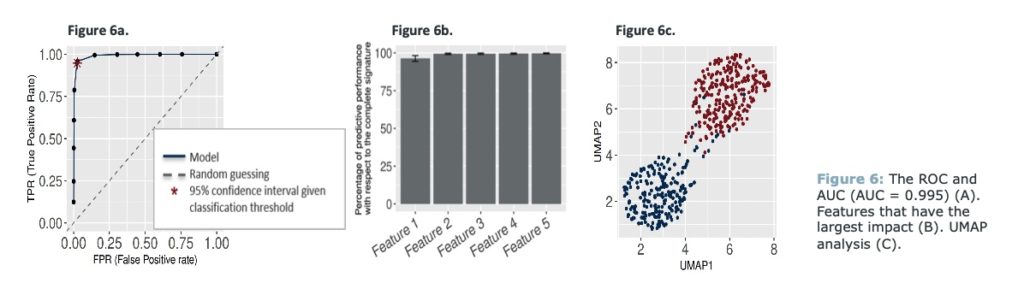
When extracting features that have the largest impact on predictive performance, features 1 and 2 mostly influenced the clustering (Figure 6b; cumulative) and a UMAP analysis based all features shows clear separation between left and right CRC (Figure 6c).
OTHER
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Stay connected to get our news first!
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Join the JADai Community!
Sign up with a FREE Basic plan! Be part of a growing community of AutoML enthusiasts
GET STARTED