Exercise and Interorgan Communication: Short-Term Exercise Training Blunts Differences in Consecutive Daily Urine ¹H-NMR Metabolomic Signatures between Physically Active and Inactive Individuals
Leon Deutsch, Biotechnical Faculty, Department of Animal Science, University of Ljubljana; Alexandros Sotiridis, Section of Sport Medicine and Biology of Exercise, School of Physical Education and Sport Science, National and Kapodistrian University of Athens,
Department of Automation, Biocybernetics and Robotics, Jožef Stefan Institute, Ljubljana; Boštjan Murovec, Faculty of Electrical Engineering, University of Ljubljana; Janez Plavec, NMR Center, National Institute of Chemistry, Ljubljana; Igor Mekjavic, Department of Automation, Biocybernetics and Robotics, Jožef Stefan Institute, Ljubljana; Tadej Debevec, Department of Automation, Biocybernetics and Robotics, Jožef Stefan Institute, Ljubljana, Faculty of Sports, University of Ljubljana, Ljubljana; Blaž Stres, Biotechnical Faculty, Department of Animal Science, University of Ljubljana, Department of Automation, Biocybernetics and Robotics, Jožef Stefan Institute, Ljubljana, Faculty of Civil and Geodetic Engineering, Institute of Sanitary Engineering, University of Ljubljana
Digital Library: https://www.mdpi.com/2218-1989/12/6/473
Abstract
Physical inactivity is a worldwide health problem, an important risk for global mortality and is associated with chronic noncommunicable diseases. The aim of this study was to explore the differences in systemic urine ¹H-NMR metabolomes between physically active and inactive healthy young males enrolled in the X-Adapt project in response to controlled exercise (before and after the 3-day exercise testing and 10-day training protocol) in normoxic (21% O2), normobaric (~1000 hPa) and normal-temperature (23 °C) conditions at 1 h of 50% maximal pedaling power output (Wpeak) per day.
Methods: Interrogation of the exercise database established from past X-Adapt results showed that significant multivariate differences existed in physiological traits between trained and untrained groups before and after training sessions and were mirrored in significant differences in urine pH, salinity, total dissolved solids and conductivity. Cholate, tartrate, cadaverine, lysine and N6-acetyllisine were the most important metabolites distinguishing trained and untrained groups.
Results
The relatively little effort of 1 h 50% Wpeak per day invested by the untrained effectively modified their resting urine metabolome into one indistinguishable from the trained group, which hence provides a good basis for the planning of future recommendations for health maintenance in adults, irrespective of the starting fitness value. Finally, the 3-day sessions of morning urine samples represent a good candidate biological matrix for future delineations of active and inactive lifestyles detecting differences unobservable by single-day sampling due to day-to-day variability.
Conclusions: The maintenance of systemic homeostasis and the response to nutritional and environmental challenges require the coordination of multiple organs and tissues. To respond to various metabolic demands, the human body integrates and builds upon a system of interorgan communication through which one tissue can affect metabolic pathways in a distant tissue. Dysregulation of these lines of communication through lack of exercise (sedentary lifestyle) and of highly energetic diets contribute to human pathologies, including obesity, diabetes, liver disease and atherosclerosis. Increasing exercise levels in the untrained apparently has the capacity to significantly reconstitute the interorgan communication towards the levels observed in the healthy trained cohort. In addition, recent technical advances such as data-driven bioinformatics on layers of information (microbiome, proteome, metabolome) expanded our understanding of the complexity of systemic metabolic crosstalk and its underlying mechanisms.
How was JADBio used?
In addition to multivariate analyses, extensive machine-learning modeling using Just Add Data Bio (JADBio) was performed to investigate the importance of metabolites and physicochemical parameters in urine samples. A total of 181,020 models were trained using extensive tuning effort. The most interpretable model was logistic ridge regression with the penalty hyperparameter lambda of 10−4 and an area under the curve (AUC) value of 0.748. In addition to AUC (Figure 6a); all other thresholds were also statistically significantly different from baseline. Data were preprocessed and standardized by imputation of means and removal of constants. Features were selected based on the test-budgeted statistically equivalent signature (SES) algorithm with the following hyperparameters: maxK = 2, alpha = 0.1, and budget = 3 × nvars. PCA plot (Figure 6b) shows that differentiation based on modeled data is not complete, which means that larger groups should be formed in the future. 12 metabolites and pH were selected as the most important features for distinguishing the trained from the untrained group based on urine. Table S2 lists all the important metabolites. The major metabolite selected by JADBio was tartrate. The power of the model obtained by using only tartrate was 73.8% (with 95% CI from 69.9% to 77.6%) (Figure S4). We applied the trained model to the test portion of our data (30% of our total dataset) and achieved validation performance with an AUC of 0.647.
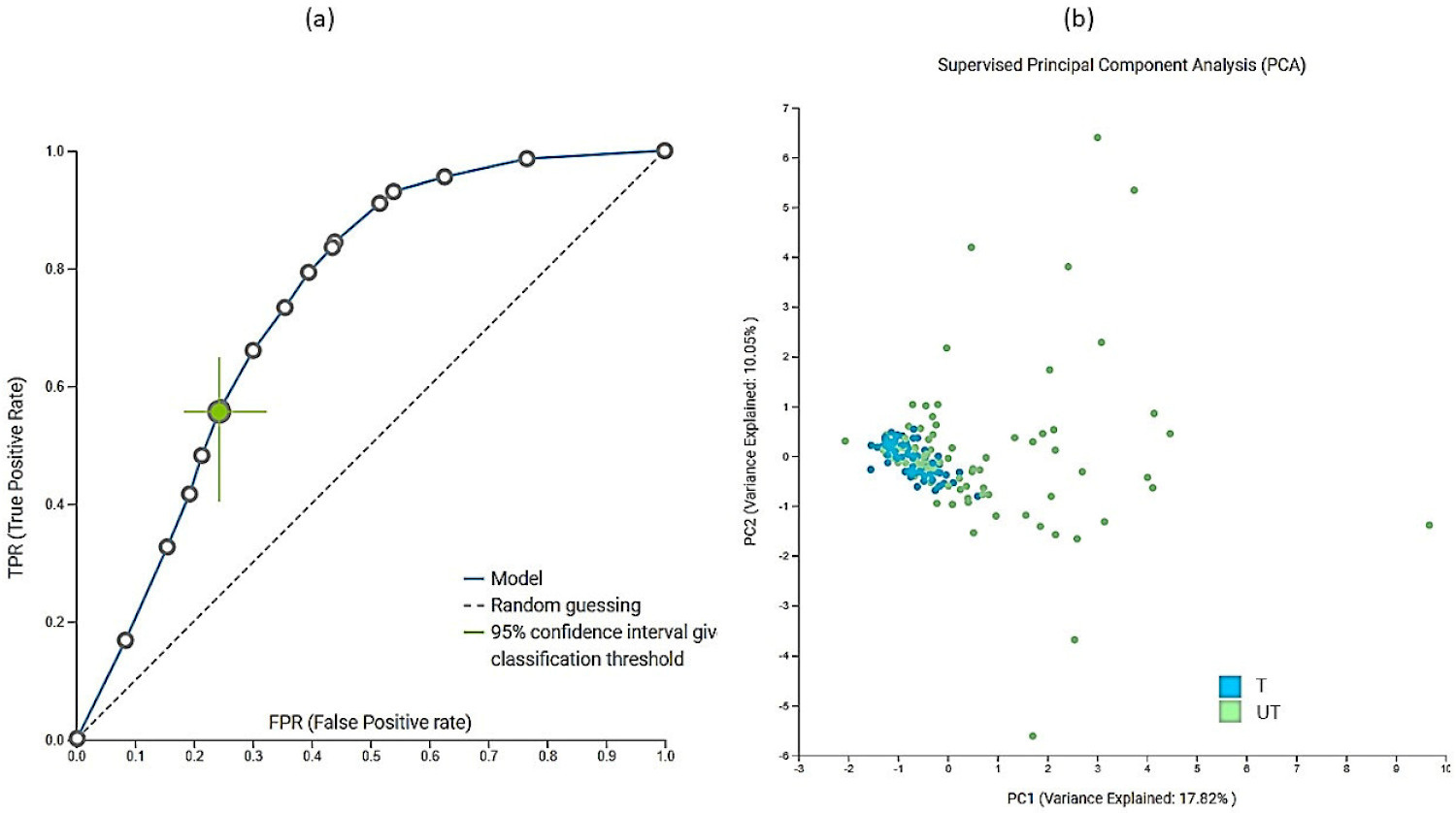
The receiver operator curve (ROC) (a) plot and PCA plot (b) of modeled data. The dimensionality reduction was performed within JADBio on a subset of the original dataset, keeping only the features included in the first signature. Features were standardized with statistical normalization ((x − µ)/σ). A total of 155 samples were included in this analysis for training the model from the entire dataset. A total of 72 samples belonged to trained and 82 samples belonged to untrained group. (Figure 6)
The main objective of the X-Adapt project was to determine the metabolic differences between trained and untrained individuals and the effects of 10 days of training on metabolism utilizing urinary metabolomics.
The resulting spectra from Bruker Avance NEO 600 MHz spectrometer were consequently analyzed using targeted quantitative metabolomics. For the latter, all spectra were randomly ordered for spectral fitting using the ChenomX profiler and the Human Metabolome Database (https://hmdb.ca/ (accessed on 24 April 2022)) compound names were used [80]. An ensemble approach to data analysis was utilized, employing three different approaches to asymmetric sparse matrix data analysis, establishing significant differences between tested groups as follows: nonparametric MANOVA (PERMANOVA) [83], MetaboAnalyst [20,21,22], and JADBio [23]. Heatmap of measured physiological parameters was generated using gplots R package. Data were normalized with scale function.
JADBio 1.4.0 with extensive tuning effort and 6 CPU was used to model various dataset selections next to the overall 336 metabolites observed in urine samples in all groups (trained vs. untrained) by splitting the total urine metabolite data into a training set and a test set in a 70:30 ratio. The training set was used for model training and the test set was used for model evaluation. The resulting model can be obtained as part of Supplementary Material (File S2) and run with Java executor for the classification of novel urine samples based on 1H-NMR in further explorations.
Electronic Supplementary Materials (Table S2, Figure S4, File S2) are available online at https://www.mdpi.com/article/10.3390/metabo12060473/s1.
OTHER
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Stay connected to get our news first!
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Join the JADai Community!
Sign up with a FREE Basic plan! Be part of a growing community of AutoML enthusiasts
GET STARTED



