This website uses cookies to improve your experience. We'll assume you're ok with this, but you can opt-out if you wish. Read More
A machine learning approach utilizing DNA methylation as an accurate classifier of COVID-19 disease severity
Scott Bowler, Division of Infectious Diseases, Department of Medicine, Weill Cornell Medicine; Georgios Papoutsoglou, Aristides Karanikas, JADBio – Gnosis DA S.A, Science and Technology Park of Crete; Ioannis Tsamardinos, JADBio – Gnosis DA S.A, Science and Technology Park of Crete, Department of Computer Science, University of Crete; Michael J. Corley, Division of Infectious Diseases, Department of Medicine, Weill Cornell Medicine; Lishomwa C. Ndhlovu, Division of Infectious Diseases, Department of Medicine, Weill Cornell Medicine
Digital Library: https://www.nature.com/articles/s41598-022-22201-4
Abstract
Since the onset of the COVID-19 pandemic, increasing cases with variable outcomes continue globally because of variants and despite vaccines and therapies. There is a need to identify at-risk individuals early that would benefit from timely medical interventions. DNA methylation provides an opportunity to identify an epigenetic signature of individuals at increased risk. Machine learning was utilized to identify DNA methylation signatures of COVID-19 disease from data available through NCBI Gene Expression Omnibus. A training cohort of 460 individuals (164 COVID-19-infected and 296 non-infected) and an external validation dataset of 128 individuals (102 COVID-19-infected and 26 non-COVID-associated pneumonia) were reanalyzed. Data was processed using ChAMP and beta values were logit transformed. The JADBio AutoML platform was leveraged to identify a methylation signature associated with severe COVID-19 disease.
Methods: Genome-wide DNA methylation of SARS-CoV-2-infected and -uninfected patients using Illumina Infinium MethylationEPIC profiling array platform from whole blood was publicly available through NCBI Gene Expression Omnibus (GEO). The training cohort (GSE16720225) consisting of 525 individuals (164 COVID-19 infected, 296 COVID-19 uninfected, and 65 with other Non-COVID-19 infections) were obtained. Participants were classified by COVID-19 severity score (SS) as follows: 0. Uninfected; 1. Released from department to home; 2. Admitted to in-patient care; 3. Progressed to ICU; and 4. Death. Severe SARS-CoV-2-infected and healthy individuals were dichotomized by SS ≥ 3 or SS = 0, respectively, resulting in a training cohort of 357 individuals. The validation cohort (GSE17481826) consisting of 102 COVID-19 infected and 26 with non-COVID-19-related pneumonia was obtained to assess the disease specificity of the model.
Where possible, raw IDAT and clinical metadata files were downloaded from GEO. IDAT files were processed using Chip Analysis Methylation Pipeline (ChAMP)27 v3.13 in R 4.1.1 following developer’s recommended pipeline using the arraytype = “EPIC” flag. In short, once loaded, samples are filtered for low quality, low bead count, presence of non-CpG probes, SNP-related probes, multi-hit probes, and probes located on X or Y chromosomes. Quality control, followed by normalization, resulted in over 850,000 methylation sites per sample for analysis. While β values are simpler to interpret, M-values have been shown to be more statistically valid for algorithm-based analysis of methylation levels28,29; thus CpGTools30 v0.10.0 in Python 3.8.8 was utilized to perform this transformation.
Results
A random forest classification model was identified from 4 unique methylation sites with the power to discern individuals with severe COVID-19 disease. The average area under the curve of receiver operator characteristic (AUC-ROC) of the model was 0.933 and the average area under the precision-recall curve (AUC-PRC) was 0.965. When applied to external validation, this model produced an AUC-ROC of 0.898 and an AUC-PRC of 0.864. These results further our understanding of the utility of DNA methylation in COVID-19 disease pathology and serve as a platform to inform future COVID-19 related studies.
Conclusions: Analysis revealed a 4 CpG methylation signature with the power to classify individuals with COVID-19 disease by leveraging JADBio AutoML platform. These findings enhance our understanding of COVID-19 disease at the methylation level and may serve to provide guidance for future COVID-19-related studies.
How was JADBio used?
Model training and feature selection
To identify a DNAm signature of severe COVID-19 disease, individuals with mild Covid disease (MCV group) were removed from the training cohort. JADBio selected a configuration where the feature selection step was performed by LASSO feature selection (penalty = 1.5) and the modeling step by a classification random forest with 100 trees and with Deviance splitting criterion (minimum leaf size = 3). Supplementary Materials include a textual summary of all analysis details along with the tested configuration and their performances. LASSO identified 4 unique methylation sites (summarized in Table 2) that lead to optimal predictive performance, namely cg17114584, cg07878065, cg03753191, and cg10778971, presented in order of importance. JADBio is also capable of identifying signatures that lead to models with statistically equivalent performance. However, in this case there were none, hence, these sites cannot be substituted with others and still obtain an equally good predictive performance (Fig. 1A). Some CpG sites may not seem to add predictive performance to the model; however, LASSO includes them as an effort to make the final model more robust to noise. Hence, there may still be some redundancy in the selected signature. By removing an individual site from the model, we observed a reduction in the performance relative to the performance achieved to the complete model (Fig. 1B). Uniform Manifold Approximation and Projection (UMAP) is a dimensionality reduction method based on manifold learning techniques and has been reported to be superior to PCA. When the selected CpG site M-values were examined with UMAP (Fig. 1C) clustering can be observed. The predictive power of our model was assessed by contrasting the cross-validated predicted probability of belonging to a specific class against the actual class of the samples (Fig. 1D).
Feature selection and training performance: The selected features and performance characteristics of the training model are summarized. COVID-19 disease = “Cov”; Healthy individuals = “Non” (Figure 1A,B,C,D)
The selected features consist of a single subset signature comprising 4 CpG sites shown by predictive performance increase when included in the model. (Figure 1A)
The predictive power losses by the model are reported for each CpG site removed. For some features, there may not be a noticeable loss in predictive power, however these features are included as an effort to make the final model more robust to noise (Figure 1B)
Uniform manifold approximation and projection (UMAP) two-dimensional space projection with LASSO feature-selected CpG methylation M-values (Figure 1C)
Separation of the predictions of the classes achieved by the model is shown in the density plot. These are the out-of-sample predictions made by the model produced by the same configuration as the final model when used for testing (e.g., during cross-validation) and not used to train the model. Well-performing models will display peaks at or close to 1 for COVID-19-infected individuals and 0 for uninfected individuals. (Figure 1D)
Model performance (training).
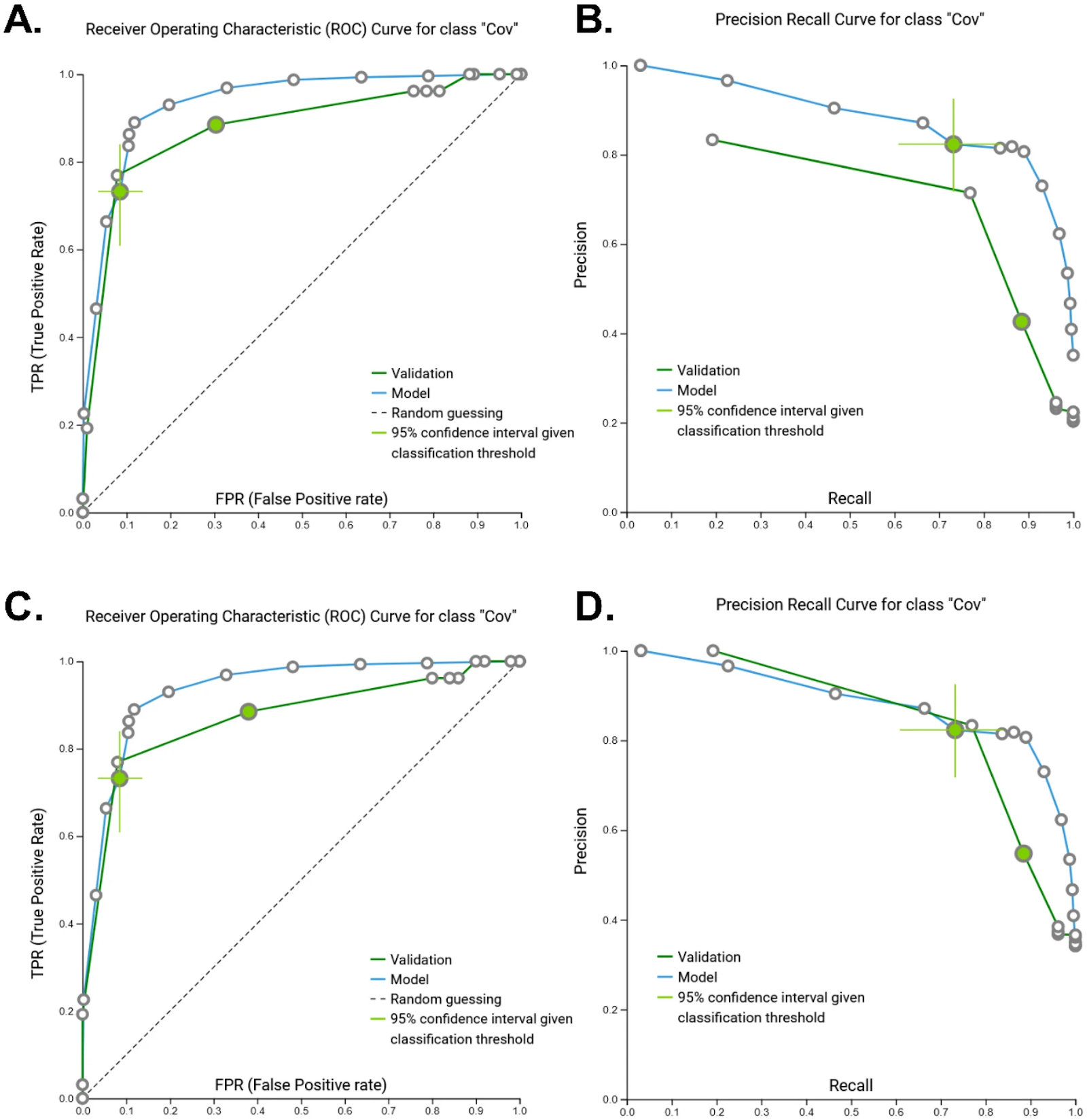
Receiver-operating characteristic (ROC) and Precision-Recall curves (PRC) for the performance of the artificial intelligence-based classification model to identify patients with severe COVID-19 disease in the training (GSE167202) cohort. AUC area under the curve; “Cov” COVID-19 disease classification. Colored circle represents the optimized classification threshold to predict COVID-19 disease, while bars extending from the circle represent 95% confidence. (Figure 2 A,B) ROC (AUC = 0.933 [0.855, 0.970]) and PRC (AUC = 0.965 [0.932, 0.986]) for model training dataset.
Receiver Operating Characteristic (ROC) Curve for class “Cov” (Figure 2A)
Precision Recall Curve for class “Cov” (Figure 2B)
External validation of the COVID-19 model
When applied to external validation data, the model produced an AUC-ROC of 0.901 with an AUC-PRC of 0.748 (Fig. 3A,B). Despite the model’s high performance classifying mild COVID-19 diseased participants within the training set, non-ICU COVID-19 patients were removed from the validation dataset to explore if the model was more precise at classifying these severe COVID-19 patients. Under these conditions, the model produced a slightly lower AUC-ROC of 0.898 while gaining in precision-recall AUC (AUC-PRC = 0.864, Fig. 3C,D) suggesting that it is more adequately trained at detecting severe COVID-19 cases than mild. Interestingly, the model was successful at identifying both healthy controls and hospitalized individuals without SARS-CoV-2 infection-associated pneumonia from those suffering from COVID-19 suggesting it may be unique to this condition.
Model performance (external validation). Receiver-operating characteristic (ROC) and Precision-Recall curves (PRC) for the validation of the artificial intelligence-based classification model to identify patients with COVID-19 disease. AUC area under the curve; “Cov” COVID-19 disease classification. Colored circle represents the optimized classification threshold to predict COVID-19 disease, while bars extending from the circle represent 95% confidence. (Figure 3A,B) AUC-ROC = 0.901 and AUC-PRC = 0.965 for the complete external validation dataset. (Figure 3C,D) AUC-ROC = 0.898 and AUC-PRC = 0.864 when non-ICU COVID-19 patients were removed from the external validation dataset.
Read more at Nature – Scientific Reports: A machine learning approach utilizing DNA methylation as an accurate classifier of COVID-19 disease severity
OTHER
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Stay connected to get our news first!
Do you have questions?
JADBio can meet your needs. Ask one of our experts for an interactive demo.
Join the JADai Community!
Sign up with a FREE Basic plan! Be part of a growing community of AutoML enthusiasts
GET STARTED